BioSift: A Dataset for Filtering Biomedical Abstracts for Drug Repurposing and Clinical Meta-Analysis
Abstract
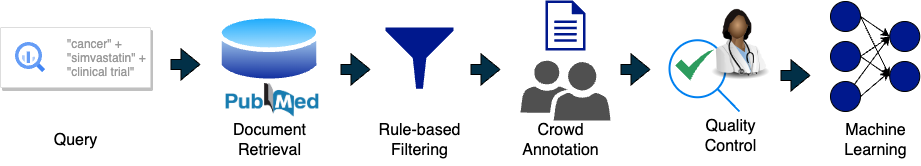
This work presents a new, original document classification dataset, BioSift, to expedite the initial selection and labeling of studies for drug repurposing. The dataset consists of 10,000 human-annotated abstracts from scientific articles in PubMed. Each abstract is labeled with up to eight attributes necessary to perform meta-analysis utilizing the popular patient-intervention-comparator-outcome (PICO) method: has human subjects, is clinical trial/cohort, has population size, has target disease, has study drug, has comparator group, has a quantitative outcome, and an “aggregate” label. Each abstract was annotated by 3 different annotators (i.e., biomedical students) and randomly sampled abstracts were reviewed by senior annotators to ensure quality. Data statistics such as reviewer agreement, label co-occurrence, and confidence are shown. Robust benchmark results illustrate neither PubMed advanced filters nor state-of-the-art document classification schemes (e.g., active learning, weak supervision, full supervision) can efficiently replace human annotation. In short, BioSift is a pivotal but challenging document classification task to expedite drug repurposing. The full annotated dataset is publicly available and enables research development of algorithms for document classification that enhance drug repurposing.
Materials
BibTeX
@inproceedings{kartchner2023biosift,
author = {Kartchner, David and Al-Hussaini, Irfan and Turner, Haydn and Deng, Jennifer and Lohiya, Shugham and Bathala, Prasanth and Mitchell, Cassie},
title = {BioSift: A Dataset for Filtering Biomedical Abstracts for Drug Repurposing and Clinical Meta-Analysis},
year = {2023},
maintitle = {SIGIR},
booktitle = {46th International ACM SIGIR Conference on Research and Development in Information Retrieval},
}

 Irfan Al-Hussaini
Irfan Al-Hussaini